

czi files) can be imported into ImageJ by using the File > Import > Bioformats command (Fiji has Bioformats installed by default). Proprietary image formats (such as the Leica. TIF hyperstack, proprietary formats such as Leica. TIF files) where each image is a single z-plane in a single color or as a single file containing all images in the stack (e.g. Depending on the type of instrument and settings used during acquisition, the image may be a sequence of individual images (typically as. Three-dimensional confocal imaging results in a collection of images as planes incrementing along the Z axis, known as an “image stack” (also known as a “z-stack”). Loading and Processing Image Stacks in ImageJ (or Fiji) In conjunction with fluorescent labeling techniques, tissue clearing dramatically increases the depths at which confocal microscopy can image into tissue, allowing for large scale reconstructions of tissue histology and morphology on large and thick pieces of tissue (> 1 mm). Visikol HISTO, CLARITY, 3DISCO) can by employed to render tissue specimens optically transparent. To increase the achievable depth at which confocal microscopes can be utilized to obtain images within tissues, clearing techniques (e.g. For deep tissue imaging (> 2mm), the use of glycerol or water immersion objectives is preferred. For most applications, air objectives work effectively the highest numerical aperture (NA) available at the magnification required should be selected, as higher NA results in more light entering the objective, which yields higher contrast and signal.

The selection of microscope objective for imaging can have serious impacts on quality of images obtained. This is commonly known as “optical sectioning” wherein thick tissue sections are employed and utilizing the optical ability of the confocal microscope as an optical “microtome,” structural details of the tissue can be revealed in three-dimensions.
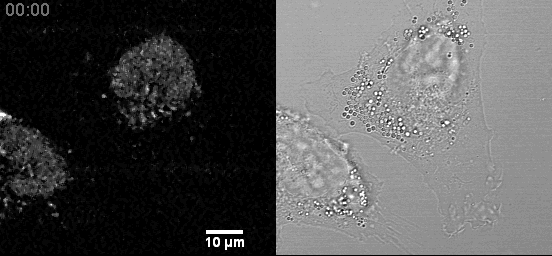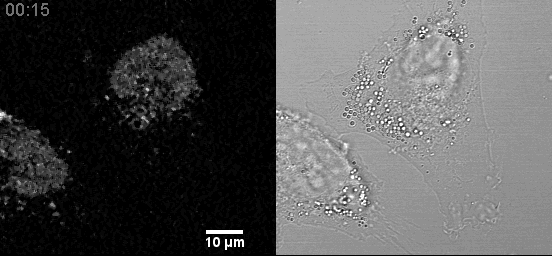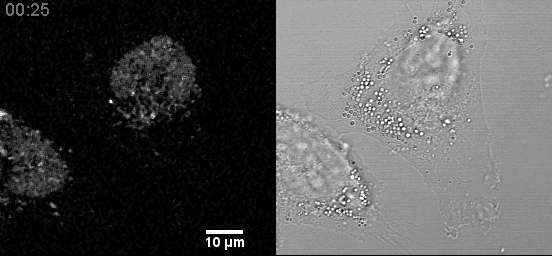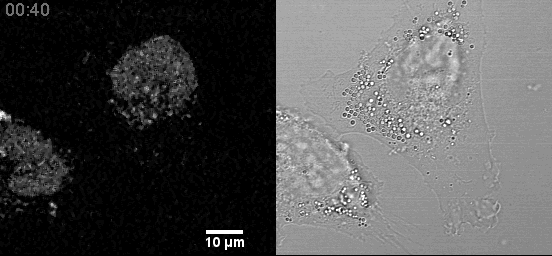
In a fixed position, providing a three-dimensional sampling of the tissue being examined. Comparison to Traditional Plant Tissue Clearing.

Immune Cell Population Analysis for Immuno-oncology.Biomarker Quantification and Colocalization.Highly-Multiplexed Immunohistochemistry Services.Cleared Tissue 3D Imaging and Analysis Services.Antibody Penetration and Pharmacokinetics.In Vitro and 3D Cell Culture Assay Services.
CZI FILE IMAGEJ SERIES
To mix those together in one series fells strange to me. This might be a bug.įrom a user point of view the additional option to open an label images using a distinct way would be more more convenient in my opinion, because the user either wants to see the real image data or the attachment. Will the label image always be the last inside the whole series?Īnd currently this label image (BioFormats 5.2.4) returns an empty image with the wrong XY dimensions.


 0 kommentar(er)
0 kommentar(er)
